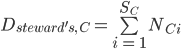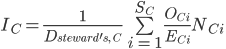Background
Introduction
Primary species occurrence records as mediated through GBIF are essential for a detailed understanding of the distribution of biodiversity in space and time. Yet, despite an impressive recent growth and now hundreds of millions of accessible records, the ability of this vast information to represent biodiversity and its change has remained largely unquantified. In the case of a relatively small and species-poor region, a small number of records may effectively characterize the distribution of species year after year. In contrast, in the case of a large and hyperdiverse region, even a large number of records may poorly represent biodiversity, especially if data is repeatedly coming from just a small portion of species, locations, or years.
Consequently, assessment of how well GBIF-mediated data represent species ranges over time, or how well-suited those data are for associated modeling and inference, requires information about an expectation: How well are mobilized data actually representing expected taxonomic, spatial, and temporal variation in biodiversity? Research by members of our team developed the use of expert-based information for this purpose (Meyer et al (2015)[2]).
We present a suite of novel informatics tools and services that build on these concepts and report on spatial and temporal data gaps and biases among countries (Oliver et al. 2021). The concepts behind our metrics have been endorsed as sound quantification of species occurrence data gaps (“Species Information Index”) by GEO BON , and recently IPBES and the CBD. We designed these tools with components embeddable in partner websites and with the potential of automated country reports that can be built out to update near-real-time.
Information about gaps and biases in this GBIF-mediated data coverage, and how successful countries and their institutions are in addressing them is vital. It can help identify priority targets for data collection and mobilization.
Metrics
Species Information Index (SII)
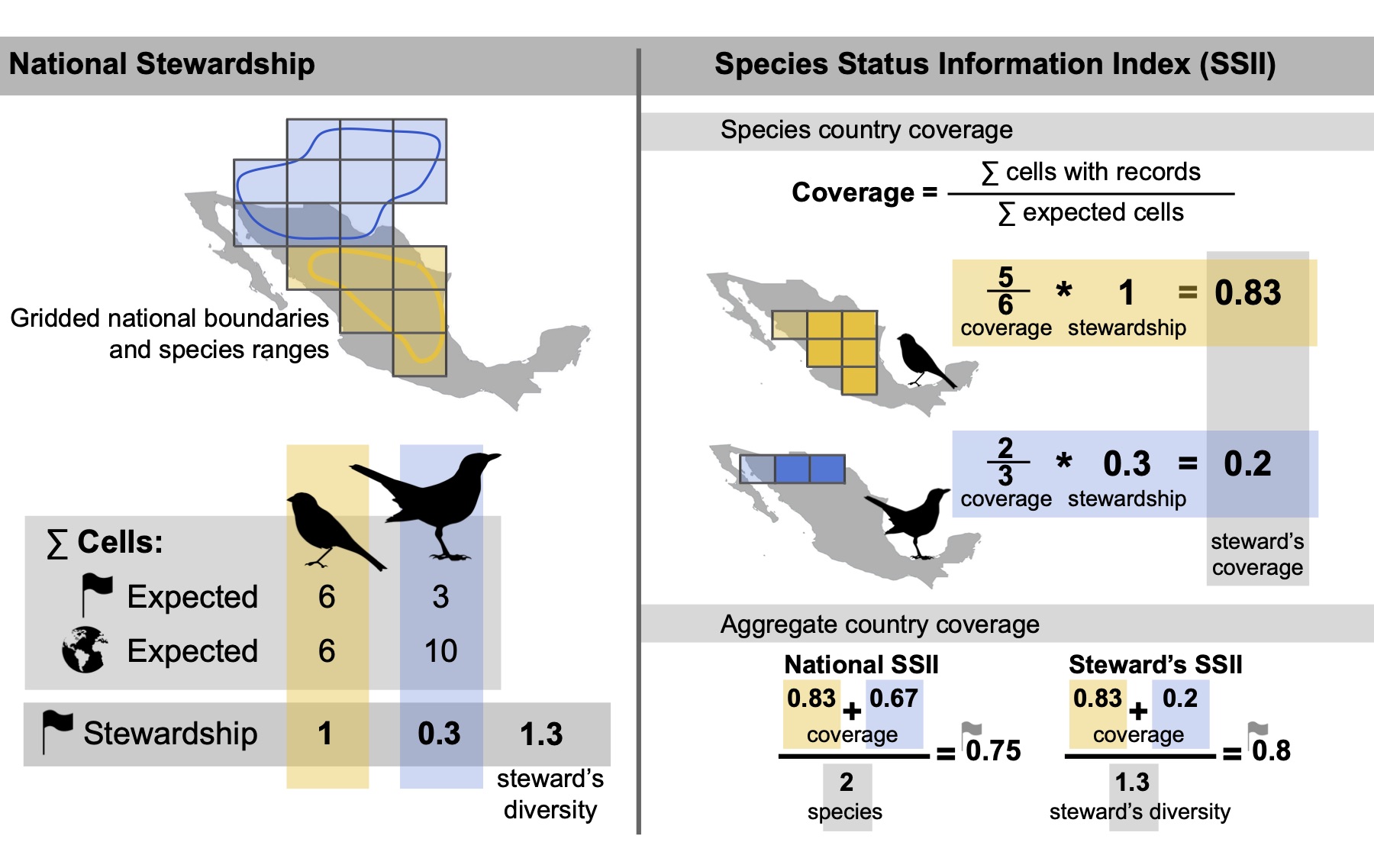
For a given species, the Species Information Index (SII) captures how well existing data covers the species’ expected range. At the species level, the SII can be computed across the entirety of the species’ expected range, ignoring national boundaries, or separately within each nation where it is expected to occur. The global SII,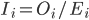 , for species i across its entire range is the proportion of expected cells, Ei, with records over a given timespan, Oi.
, for species i across its entire range is the proportion of expected cells, Ei, with records over a given timespan, Oi.
At the national level, for a given taxon with Sc species expected in country C, we define the SII, IC, as follows, distinguishing two formulations:
- National SII: This index measures how well, on average, species are documented in a given nation over a given timespane, in this case per year. The index value I for country C in a particular year is given by the arithmetic mean among expected species Sc of the proportion of expected cells in country C, Ec, with records from that year, Oc (Fig. 1b):
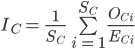
- Steward’s SII: This index adjusts the national coverage based on nations’ stewardship of species, upweighting the documentation of species for which a nation has particularly high stewardship. National species stewardship
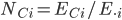 defined as the proportion of global cells that country C holds for a species i where
defined as the proportion of global cells that country C holds for a species i where 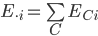 is the total number of expected grid cells for a species across all countries (Fig. 1, top panel). National species stewardship can then be used to weight both the national-level coverage, Ic, and number of species that country C is responsible to document, Dsteward's,C (Fig. 1c):
is the total number of expected grid cells for a species across all countries (Fig. 1, top panel). National species stewardship can then be used to weight both the national-level coverage, Ic, and number of species that country C is responsible to document, Dsteward's,C (Fig. 1c): 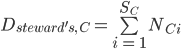
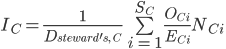
Calculations
Calculations are performed over an equal area global grid of ca. 110-km resolution or which expert expectations are deemed broadly reliable (see Hurlbert & Jetz (2007) [1] ). Expert expectation information is provided expert-based range maps (see Sources), where the sources used are assumed to be broadly characteristic for the past 35 years. We carefully developed synonym lists to match species names in GBIF-mediated data to names used for the expert information. We note, however, several caveats: expert maps may suffer from errors of omission and commission that even at this coarse spatial resolution may slightly affect metric values; taxonomic mismatches may remain in the data. In future updates, we plan to replace expert maps with increasingly sophisticated multi-source, modeled maps, and further improve the synonym match-up.
Taxonomic harmonization
To prevent missing or double-counting species due to synonymy, all scientific names in GBIF and range map datasets are harmonized under common taxon concepts (see Sources). For this purpose, we carefully developed synonym lists to match species names in GBIF-mediated data to names used for the expert information. For each major species group, we selected a well-curated taxonomic database as our ‘master taxonomy’, that defines accepted species delimitations. To each accepted species name, we linked additional scientific names that are fully or partly included in the respective taxon concept, including synonyms, subspecies, and typographical variants and spelling mistakes. To be able to interpret all scientific names in range map and GBIF datasets, we integrated our master taxonomies with various additional sources of scientific names. Any remaining non-matching names were matched using approximate string matching and afterwards individually validated.
Scientific name resolution
The presented indicators refer to native biota only. To validate records geographically, we thus excluded any records that fell outside of the respective species’ known extents of native occurrence (as inferred from gridded range maps). A major problem with records in aggregative databases is that the originally intended taxon concepts behind ambiguous scientific names (such as many pro parte synonyms) are typically unknown. However, these can often be inferred indirectly from records’ geographical locations. Here, we do so by inferring taxonomic identities of ambiguously named records through spatial overlays with the range maps of all accepted species to which these names could potentially refer. In cases where ambiguously named records overlap with ranges of more than one ‘candidate’ accepted species, we assume that these names reflect the taxon concepts in our master taxonomy. If ambiguities still persist, we assume that all records of a given ambiguous name in a given grid cell refer to only one of the candidate accepted species, hence our completeness index may be slightly conservative in these cases. For more information, see Sources .
References
- Hurlbert, A. H., and W. Jetz. 2007. Species richness, hotspots, and the scale dependence of range maps in ecology and conservation. PNAS 104:13384-13389)
- Meyer, C., H. Kreft, R. Guralnick, and W. Jetz. 2015. Global priorities for an effective information basis of biodiversity distributions. Nature communications 6: 8221
- Oliver, R. Y., Meyer, C., Ranipeta, A., Winner, K., & Jetz, W. (2021). Global and national trends, gaps, and opportunities in documenting and monitoring species distributions [Data set]. PLOS Biology. https://doi.org/10.48600/MOL-3Y3Z-DW77
- Oliver, R.Y., Meyer, C., Ranipeta, A., Winner, K., Jetz, W. (2021) Global and national trends, gaps, and opportunities in documenting and monitoring species distributions. PLoS Biology 19(8): e3001336. doi.org/10.1371/journal.pbio.3001336

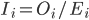 , for species i across its entire range is the proportion of expected cells, Ei, with records over a given timespan, Oi.
, for species i across its entire range is the proportion of expected cells, Ei, with records over a given timespan, Oi. 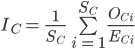
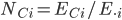 defined as the proportion of global cells that country C holds for a species i where
defined as the proportion of global cells that country C holds for a species i where 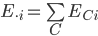 is the total number of expected grid cells for a species across all countries (Fig. 1, top panel). National species stewardship can then be used to weight both the national-level coverage, Ic, and number of species that country C is responsible to document, Dsteward's,C (Fig. 1c):
is the total number of expected grid cells for a species across all countries (Fig. 1, top panel). National species stewardship can then be used to weight both the national-level coverage, Ic, and number of species that country C is responsible to document, Dsteward's,C (Fig. 1c):